Sacrifice for the Swarm
Souvik Bhattacharyya and Rasika M Harshey explain how bacterial cells killed by antibiotics help others to avoid the same fate – a process known as ‘necrosignalling’
Feb 22nd 2021
In the animal world direct inter-species competition often involves aggression and the death of competitors. This is also true for microorganisms, which can produce antibiotic compounds and carry many arrows – both chemical and physical – in their defensive quiver.
The structure and mechanism of action of antibiotics, both natural and synthetic, are varied. Bacterial defence mechanisms to counteract the antibiotic are equally varied, and can include chemical inactivation, processes to pump the antibiotic out of the cell or the formation of biofilms to further inhibit the action of antibiotics. Bacteria can even enter a dormant state that deprives the antibiotic of active metabolic processes to ambush. Swarming is an uncommon example of collective migration seen in bacteria, where cells band together to navigate solid surfaces using appendages called flagella[1].
Swarming populations have been shown to exhibit temporary resistance to a broad class of antibiotics, known as swarm-specific resistance (SR). This adaptive resistance only occurs when the bacteria are in a swarm and dissipates when the bacteria are dispersed in liquid. Unlike biofilm formation or dormant states that resist antibiotic action by low metabolic activity, swarming is an energy-intensive process.
 A better understanding of the dynamics of swarming behaviour could help inform the treatment of drug-resistant infections.
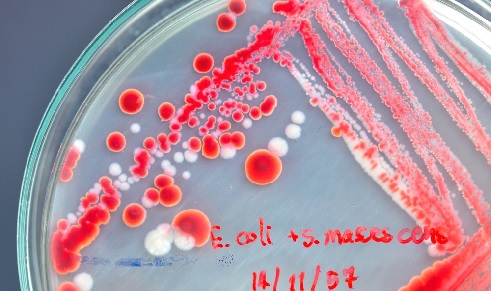
A better understanding of the dynamics of swarming behaviour could help inform the treatment of drug-resistant infections. In a study published in 2010[2], our laboratory designed a simple assay to measure SR and found approximately a third of the swarm population was being killed by the antibiotic. This raised several questions. Was this amount of death simply collateral damage or did it have any bearing on the mechanism of SR? Were the dead cells providing a physical barrier against antibiotic penetration or had they helped the remaining bacteria evade the antibiotic in some other way?
A follow-up study published in 2020[3] exploited the assay design of the 2010 study to show that the dead cells were not simply collateral damage, nor were they providing a physical barrier, but were instead contributing actively to SR by a previously undescribed mechanism. A key finding of the 2020 study was that a sub-population of cells in an Escherichia coli swarm is more susceptible to very low doses of antibiotic. When a swarm moves into antibiotic territory this population is the first to die, spilling its internal contents into the swarm milieu.
We could reproduce the SR phenomenon by simply adding the cellular contents of killed cells to the swarm without the presence of the antibiotic, so we surmised that some component of the spilled cell contents must be responsible for bestowing SR to the live swarm. Using this observation, and the assay developed in the 2010 study, we purified the active SR ingredient and identified it as a protein called AcrA. We were astonished to find that this protein is an internal part of a specific efflux pump, AcrAB-TolC.
E. coli encodes multiple such pumps, which normally open for business when antibiotics enter the cell and interact with the cytoplasmic side of the pump. Even more surprising was that AcrA, when released from dead cells, binds to the external component found in many efflux pumps, activating them to start pumping out the antibiotic. In other words AcrA was mimicking the cellular response seen when antibiotics enter live cells – except it was cranking the pump from the outside, helping cells to pump the antibiotic out before it before it caused damage. We called this phenomenon ‘necrosignalling’. In the four bacterial species tested we found necrosignalling to be conspecific.
A curious aspect of necrosignalling is that it is only manifested in the swarm, which by definition is a collective group. That a subset of the group is alarm-calling the larger group by ‘sacrificing’ itself could be seen as akin to conventional altruism in evolutionary biology, where an organism is said to behave altruistically when its behaviour benefits other organisms at a cost to itself.
Altruistic behaviour is particularly common in species with complex social structures. The conspecificity of necrosignalling we have observed strengthens the case that this kind of alarm-calling is related to altruism. If necrosignalling is a feature of a bacterial societal organisation, other social phenomena such as biofilms might also harbour similar signalling mechanisms.
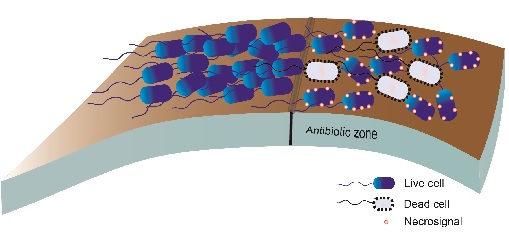 Fig.1. Necrosignalling in a population of bacteria swarming on a surface. When the swarm cells enter a zone containing antibiotics some cells die and release necrosignals that enhance resistance in other cells
Fig.1. Necrosignalling in a population of bacteria swarming on a surface. When the swarm cells enter a zone containing antibiotics some cells die and release necrosignals that enhance resistance in other cellsGenetic resistance
Antibiotics are a double-edged sword: while we need them to eliminate bacterial infections their use helps select for strains with bacterial resistance. The longer bacteria can tolerate an antibiotic while swarming, or while hunkering down as attached biofilms, the more time that population of cells has to acquire genetic resistance.
Can our knowledge of the necrosignal and other social signalling in microbes help us combat antimicrobial resistance (AMR)? We imagine two areas of antibiotic research that our study may inform. The first is in the design of new antibiotics: here, one might consider developing a suite of necrosignal inhibitors to be given in combination with antibiotics.
The second is in design of treatment regimens. As well as sensitivity tests to determine the choice of antibiotic, tests should run in parallel to determine the bacterial load, therefore swarming characteristics, of the infection. High bacterial loads would be particularly fertile grounds for generating necrosignals. Formulating and administering a lethal antibiotic dose that considers potential necrosignalling might help lower the risk of AMR during the antibiotic treatment.
Finding new antibiotics to kill pathogenic bacteria is a Sisyphean task. Scientists are throwing a wide net, exploring new niches for new organisms and new bacterial poisons only to find that the magic in the ‘magic pill’ is ephemeral. Although there have been some successes in recent years in the discovery of new classes of antibiotics – for example, teixobactin isolated from the soils of Maine in 2015[4] has a new lipid target, and lugdunin isolated from commensals in the human nose in 2016[5] is a non-ribosomally made peptide that apparently inhibits translation – there is no reason to doubt that within just a few years of introducing the new antibiotics the evolutionary arms race will reassert itself.
While we await the next big discovery in eliminating bacterial pathogens, necrosignalling could be exploited as an extra tool in our strategy – a face mask to tide us over until we have an effective cure.
Souvik Bhattacharyya is post-doctoral Fellow at the Department of Molecular Biosciences, University of Texas at Austin
Rasika M Harshey is professor at the Department of Molecular Biosciences, University of Texas at Austin
1) Harshey, R. M. Bacterial motility on a surface: many ways to a common goal. Ann. Rev. Microbiol. 57, 249–273 (2003).
2) Butler, M. T. et al. Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. USA, 107(8), 3776–3781 (2010).
3) Bhattacharyya, S. et al. Dead cells release a ‘necrosignal’ that activates antibiotic survival pathways in bacterial swarms. Nat. Commun. 11(1), 4157 (2020).
4) Ling, L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature 517(7535), 455–459 (2015).
5) Zipperer, A. et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature, 535(7613), 511–516 (2016).


