Spatialomics: Life in 3D
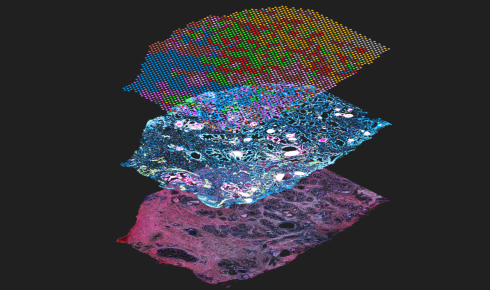
Image: A Visium CytAssist assay showing spatial gene expression (top layer) and protein expression (middle), courtesy of 10x Genomics
Spatial omics is allowing biologists to reveal the finer details of human development, particularly in the developing embryo, writes Sachin Rawat
December 11th 2023
Miscarriages are common, accounting for 10–20% of all known pregnancies, and many more pregnancies end before they are known. Early development is a delicate phenomenon that can be derailed by the slightest of errors. There are still huge gaps in what we know about this stage of development, particularly between weeks two and six.
A major reason this black box persists is the internationally recognised ethical limit of not culturing human embryos beyond the 14th day. However, developmental biologists can circumvent this limitation by studying development in other animals and, more recently, organoids and synthetic embryos. While these approaches recapitulate features of human embryos, they are considerably different.
Specifically, owing to variations in morphology and size, cells in organoids or synthetic embryos don’t always map to exactly the same positions in space as the organs and embryos that they mimic. Spatial omics, a set of emerging technologies that map various omics data to physical coordinates, is addressing this gap.
Spatial omics go beyond and complement insights from single-cell studies. For example, single-cell transcriptomics can reveal the different cell types in a tissue based on which genes are being expressed in different populations, but it doesn’t say anything about where the different cell types are. Spatial transcriptomics, on the other hand, preserves the spatial context and reveals where the genes are expressed too.
Spatially resolved development
In a developing embryo cells grow rapidly, assume new identities (including some transient cell types) and move around to set the outline for the body. “The embryo changes shape a lot,” says Naomi Moris, a developmental biologist at the Francis Crick Institute. These interactions rely on the precise location of cells at a particular point in space and time. “Understanding where the genes are expressed and how that relates to the morphology is important.”
Most prominent spatial omics techniques combine imaging and methods based on next-generation sequencing (NGS) to explore spatially resolved gene expression¹. Imaging-based spatial transcriptomics techniques include in situ hybridisation and in situ sequencing – both reveal gene expression with spatial coordinates, like pincodes on maps. Other NGS-based techniques use probes to map RNA, often in conjunction with proteins, in tissues. These probes are spatially barcoded, which means that when they detect a specific RNA or protein, we know where in the tissue it is.
With advances in high-throughput imaging biologists can now simultaneously profile dozens of different molecules at single-cell resolution with multiplexed imaging. Technical advances in spatially resolved proteomics and other omics are also under way. Bit by bit these techniques are uncovering how the spatial context of cells constrains and shapes how they move and interact, creating different niches within the developing embryo.
For example, Moris and colleagues look at embryonic stem cells and how they interact with one another to form the derivatives of the three germ layers. To study this slice of early development they use gastruloids. Spanning just under a millimetre, these are organoids wherein embryonic stem cells aggregate to resemble gastrulating embryos.
 Adding detailed spatial location data to multiple omics assays is leading to powerful new tools for developmental biology. Image courtesy of 10x Genomics.
Adding detailed spatial location data to multiple omics assays is leading to powerful new tools for developmental biology. Image courtesy of 10x Genomics.
Because of restrictions on working with human embryos there’s little human embryo data. “Having that spatial information in the data sets that we do have is quite powerful that way to go back and validate some of the models that we have [of human embryonic development],” says Moris.
Building maps and atlases
This year reports of synthetic embryos have frequently made the news², often spurring debates about whether they need greater regulation in line with human embryos. Many, including Moris, argue that they aren’t true embryos and are instead only embryo-like, without a heartbeat or beginnings of a brain. She added that there is nothing synthetic about them either, as they represent “how the cells coordinate with one another and enact their intrinsic programmes”.
While the debate continues there is no denying that these models are advancing our knowledge of early development. Various research groups and consortia are working on maps and atlases of different systems in development. These are rich repertoires of all different cell populations, placed in their spatial and temporal contexts in specific systems.
For example, the Human Cell Atlas (HCA) project has led to spatially resolved atlases of the embryonic limb³, the first-trimester brain4 and the extraembryonic yolk sac5, among other embryonic regions. Working with 30-year-old samples, a spatial omics study mapped the trophoblast landscape during implantation, revealing the trajectory of foetal cells as they invaded the maternal cells in the uterus6. The Human Developmental Cell Atlas (HDCA) initiative7, a part of the HCA, aims to build a complete atlas of all cell types during development.
Much of the phenotypic changes seen in maps and atlases are driven by interactions happening at the level of individual cells.
“Spatial information allows us to know neighbourhood relationships between cells,” says Léo Guignard, a computational biologist at the Turing Centre for Living Systems (CENTURI) in France. By applying computational methods and network theory to spatially resolved data sets, biologists can figure out which cells are next to each other and how they’re interacting.
Guignard stresses the relevance of these spatial omics-derived maps and atlases as standards to compare organoids and synthetic embryos under investigation, such as to study mechanisms of developmental disorders. “If you have an atlas of the wild type and some sort of statistical metric, you can see how different it is and what genes are missing,” says Guignard.
The power of multi-omics
On their own spatial transcriptomics and spatial proteomics capture only a slice of what’s happening in a developing embryo – and there’s now a push towards spatial ‘multi-omics’ approaches to capture the true extent of the messy dynamics of an embryo. Combining data from different omics and imaging modalities to create coherent models, however, comes with its challenges.
For instance, often different patterns show up while assessing the same sample with different omics techniques. Complicating things further, the differences could have a biological basis to them. Computational biologists are tackling these data integration challenges, broadening the applicability of spatial omics.
With synthetic embryos and organoids that mimic development more accurately, scientists hope to probe cellular dynamics further into human development. Beyond miscarriages, what they will learn will have implications for our understanding of cancers, the brain, developmental disorders and regenerative medicine. This new context- and space-specific version of transcriptomics is starting to make more traditional methods – where tissue is homogenised and analysed to yield averaged gene expression across many different cells – look rather vague.
Sachin Rawat is a freelance science and technology writer based in Bangalore. He holds a master’s degree in biotechnology.
1) Williams, C. G. et al. An introduction to spatial transcriptomics for biomedical research. Genome Med. 14, 68 (2022).
2) ‘Synthetic human embryos created in groundbreaking advance’. The Guardian, 14 Jun 2023.
3) Zhang, B. et al. A human embryonic limb cell atlas resolved in space and time. BioRxiv (2022).
4) Braun, E. et al. Comprehensive cell atlas of the first-trimester developing human brain. BioRxiv (2022).
5) Goh, I. et al. Yolk sac cell atlas reveals multiorgan functions during human early development. Science 381, eadd7564 (2023).
6) Arutyunyan, A. et al. Spatial multiomics map of trophoblast development in early pregnancy. Nature 616, 143–151 (2023).
7) Haniffa, M. et al. A roadmap for the Human Developmental Cell Atlas. Nature 597, 196–205 (2021).


